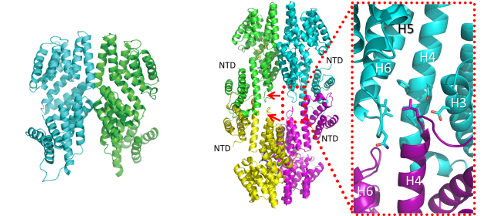

Left: Cartoon representation of the side view of the dimeric structure of apo RappLS20. Right: RappLS20 tetramerization, side view of the peptide-bound tetramer. The red arrows indicate the loops connecting helices H4 and H5. (C) Zoom of the area around the N-terminus of helix H4, showing the insertion of this helix into the opposite monomer. The homotetramerization caused by the foot-2-foot interactions of the NTDs of RappLS20 provides an explanation for the activation of the RcopLS20 partner. In the absence of the peptide, the NTDs are positioned such that they allow the interaction with RcopLS20. However, upon binding the signaling peptide, the NTDs shift outwards, facilitating the formation of the homotetramer, leading to a change of the interaction surface of the NTDs that is no longer available for interactions with RcopLS20. Right: ALBA team.
Cerdanyola del Vallès, 20th November 2020 Bacteria possess mechanisms to establish communication between cells. This is especially important in bacterial conjugation, a process that allows bacteria to share genetic material. This is often used by bacteria to transfer antibiotic resistance genes and other virulence factors to neighbor cells, increasing the antibiotic resistance spread.
Now, a research team of ALBA scientists report the structural mechanism by which two proteins, Rap and Rco, act together to regulate conjugation. Rco is a repressor of conjugation, whereas Rap binds Rco and prevents Rco-mediated conjugation repression, thus resulting in an activation of the conjugation mechanism. The main results of the study show that Rap contains a binding pocket were a short peptide can bind, producing structural changes in Rap that forces its tetramerization, releasing Rco for blocking conjugation. Tetramerization occurs through an interaction that scientists named “foot-2-foot”, which differs significantly from the model proposed for other proteins of the Rap family.
“We have demonstrated for the first time direct interaction between Rap and Rco proteins and unraveled the mechanism of how the peptide binding to Rap prevents this Rap-Rco interaction, repressing the conjugation process”, explains Isidro Crespo, researcher at ALBA.
His colleague Nerea Bernardo, scientist at ALBA too, adds that “we show that Rap protein without binding is dimeric and that peptide-bound Rap forms N-terminal domain-mediated tetramers. In addition, we report that Rap binds Rco directly in the absence of the peptide and, its addition releases Rco from Rap. This allows Rco to bind the promotor region of crucial conjugation genes blocking their expression.”
Information on the structural basis of the interaction mechanism was gathered by the combination of information from several synchrotron techniques. Macromolecular crystallography at the XALOC beamline of ALBA showed the Rap protein atomic structure and the changes induced upon peptide interaction, suggesting the tetramerization. This was confirmed by SAXS (small-angle X-ray scattering performed at NCD-SWEET of ALBA and BM29 of the ESRF), SEC (size exclusion chromatography) and AUC (analytical ultracentrifugation) studies. “This approach combining the four techniques has been crucial in the elucidation of the oligomerization behavior of Rap and its interaction partners”, commented Roeland Boer, beamline responsible of XALOC.
Finally, it was confirmed that tetramerization produced the dissociation from Rco, being freed on Rap-peptide interaction. Researchers highlight that knowing the mechanism of conjugation inhibition may help in finding clinical strategies to avoid bacterial conjugation in order to prevent antibiotic resistance spread. Results show that the addition of the peptide produces the dissociation of Rap-Rco complex, and the free Rco blocks the conjugation. So, following this strategy, drugs targeting Rap could enhance Rco conjugation blocking, preventing bacteria to spread antibiotic resistance genes or other virulence factors.
The results represent another step forward towards preventing human diseases since these proteins are found in a wide range of human commensal or pathogenic bacterial genera such us Bacillus, Streptococcus and Enterococcus.
Reference: Isidro Crespo, Nerea Bernardo, Andrés Miguel-Arribas, Praveen K Singh, Juan R Luque-Ortega, Carlos Alfonso, Marc Malfois, Wilfried J. J. Meijer, Dirk Roeland Boer. Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization. Nucleic Acids Research, 2020, 48, 14 8113–8127. DOI:10.1093/nar/gkaa540
ALBA team (Roeland, Nerea, Isidro) led the study, with help of the NCD team (Marc Malfois) and Madrid collaborators, specialists in bacterial conjugation (Andrés Miguel-Arribas, Praveen K. Singh,Juan R. Luque-Ortega, Carlos Alfonso, Wilfried J.J. Meijer).




