Cerdanyola del Vallès, 31st January 2019. Controlling the lifetime of cellular proteins is an essential mechanism for regulating their activity and the cell cycle to work properly. This cycle allows cells to divide and thus make the tissues grow and regenerate. The control of the different stages of the cell cycle is played by different types of proteins that are being replaced as the cycle progresses. The cells have developed a mechanism to mark proteins that must be destroyed to be replaced, this process called ubiquitination, and the brand is the formation of a chain with the protein ubiquitin . Proteins marked (with ubiquitin) are eliminated by a type of cell called large dump proteasome responsible for crushing them. The regulation of this process is very important for cell cycle function properly, otherwise may lead to alterations in cell proliferation processes that could lead to cancer, where tissue grows without control.
In the work that the researchers from IBB (Institute of Biotechnology and Biomedicine) of the UAB have just published in the journal Nature Communications presented the structural characterization and discovery of a new mechanism of regulation of an enzyme that cuts the chains of ubiquitin, called USP25. This enzyme removes the chains of ubiquitin proteins and has a marked impact on its function in cell from being degraded by the proteasome.
The three-dimensional structure of this enzyme, obtained through protein crystallography and ALBA Synchrotron light in the XALOC beamline, has allowed us to observe the presence of an assembly with two different types of protein quaternary structure: tetramer and dimer. And more importantly, the tetramer and dimer is idle is active. Thus alternating between the two assemblies regulates the activity of this enzyme and, indirectly, the lifetime of important cellular proteins.
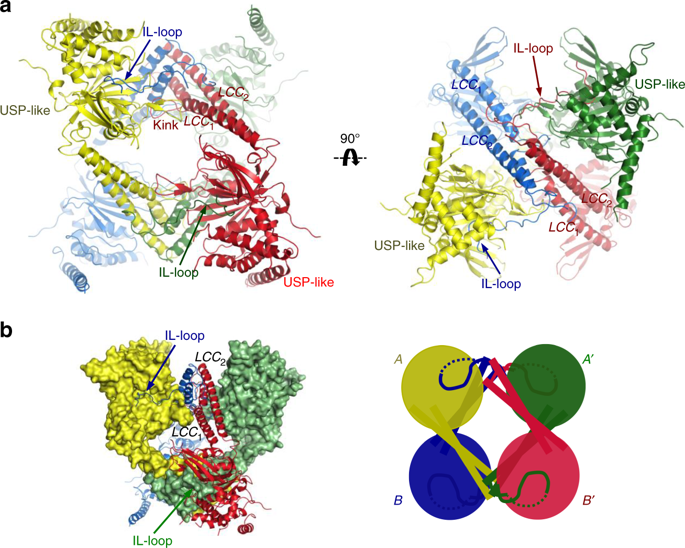
USP25 is a homotetramer formed by the assembly of two dimers. a Ribbon representation of the USP25 tetramer. Each subunit is represented with a different color. LLC and IL-loop domains are labeled. Different orientations of the USP25 tetramer. b Mixed surface and ribbon representation of the homotetramer structure of USP25. Cartoon image of the tetramer assembly formed by the assembly of two homodimers (A, A′ homodimer and B, B′ homodimer). Image from Nature Communications.
Liu, B., Surena-Gómez, M., Zhen, Y., Amador, V., Reverter, D. A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25. Nature Communications (2018 ). DOI: 10.1038 / s41467-018-07510-5.




